Research
SIGNALING TO CHROMATIN FOR RAPID TRANSCRIPTION RESPONSES
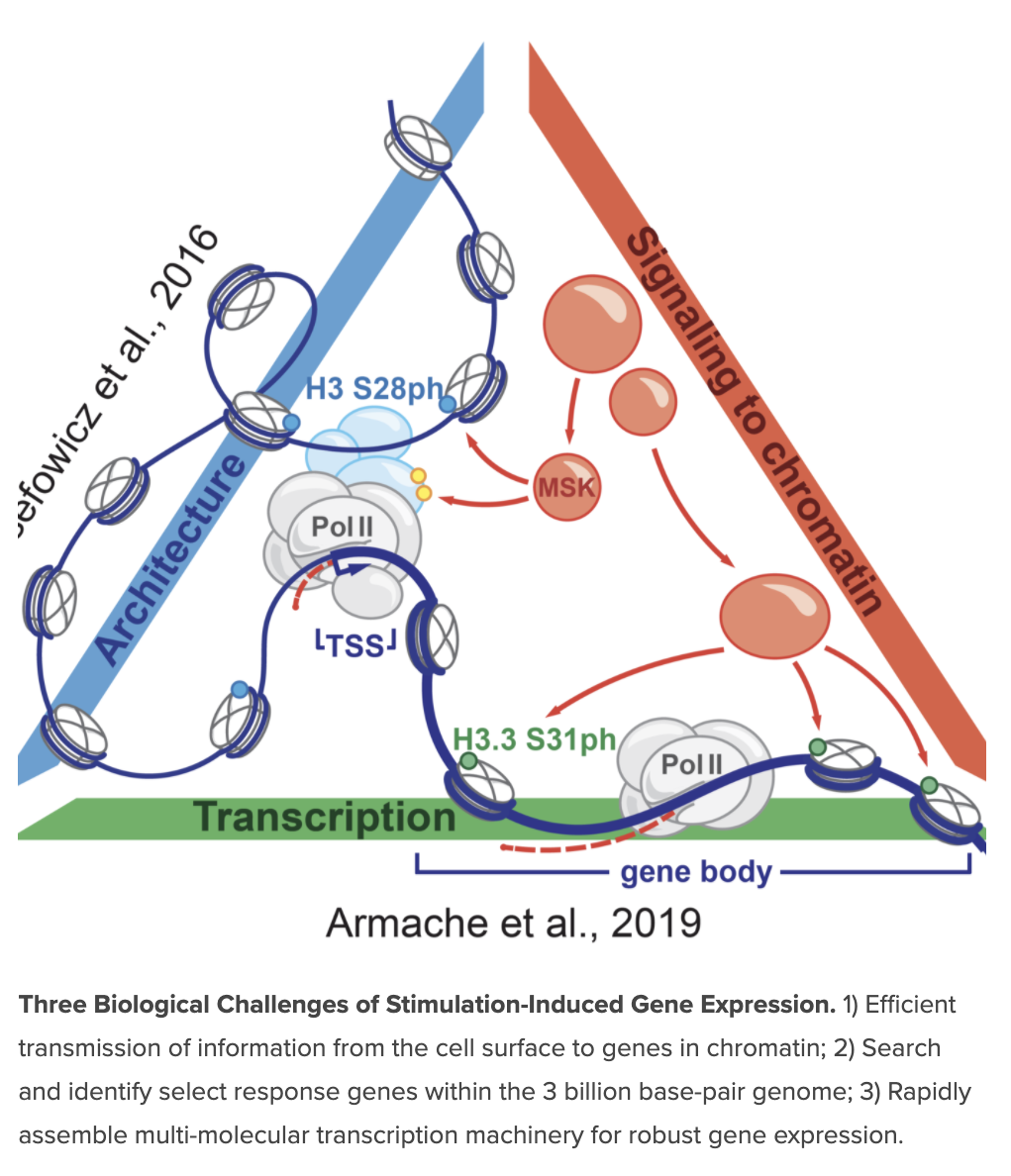
Signals are transmitted to chromatin to facilitate rapid, robust, and selective gene expression within the three billion base-pair genome in response to environmental cues, such as pathogen sensing. The goal of our ongoing research is to reveal mechanisms allowing for this scope and selectivity, and to understand them in the context of dynamic and fluid chromatin and all of its constituents.
We have found that kinases downstream of environmental signaling pathways activate both transcription factors and histones in chromatin to synergistically drive rapid, high-level transcription of stimulation induced genes. These “signaling to chromatin” pathways are notable in their broad utilization in diverse rapid cellular responses, and also by their constitutive activation (potential requirement?) in many cancers and inflammatory disease. A central hypothesis is that there exists a class of dedicated epigenetic mechanisms, histone modifications, and regulatory factors specific for selective stimulation-induced transcription to drive de novo, high-level gene expression even while 1000s of other genes are constitutively expressed. These mechanisms critically enable rapid, tailored cellular responses but may be dysregulated in disease.
Anja Armache et al.2020
Histone H3.3 phosphorylation amplifies stimulation-induced transcription
Nat. doi: https://doi.org/10.1038/s41586-020-2533-0 [pubmed]
Josefowicz SZ, Shimada M, Armache A, Li CH, Miller RM, Lin S, Yang A, Dill BD, Molina H, Park H-S et al. 2016.
Chromatin Kinases Act on Transcription Factors and Histone Tails in Regulation of Inducible Transcription.
Mol Cell. 64(2):347-361. [pubmed]
MOUSE MODELS FOR FUNCTIONAL HISTONE GENETICS
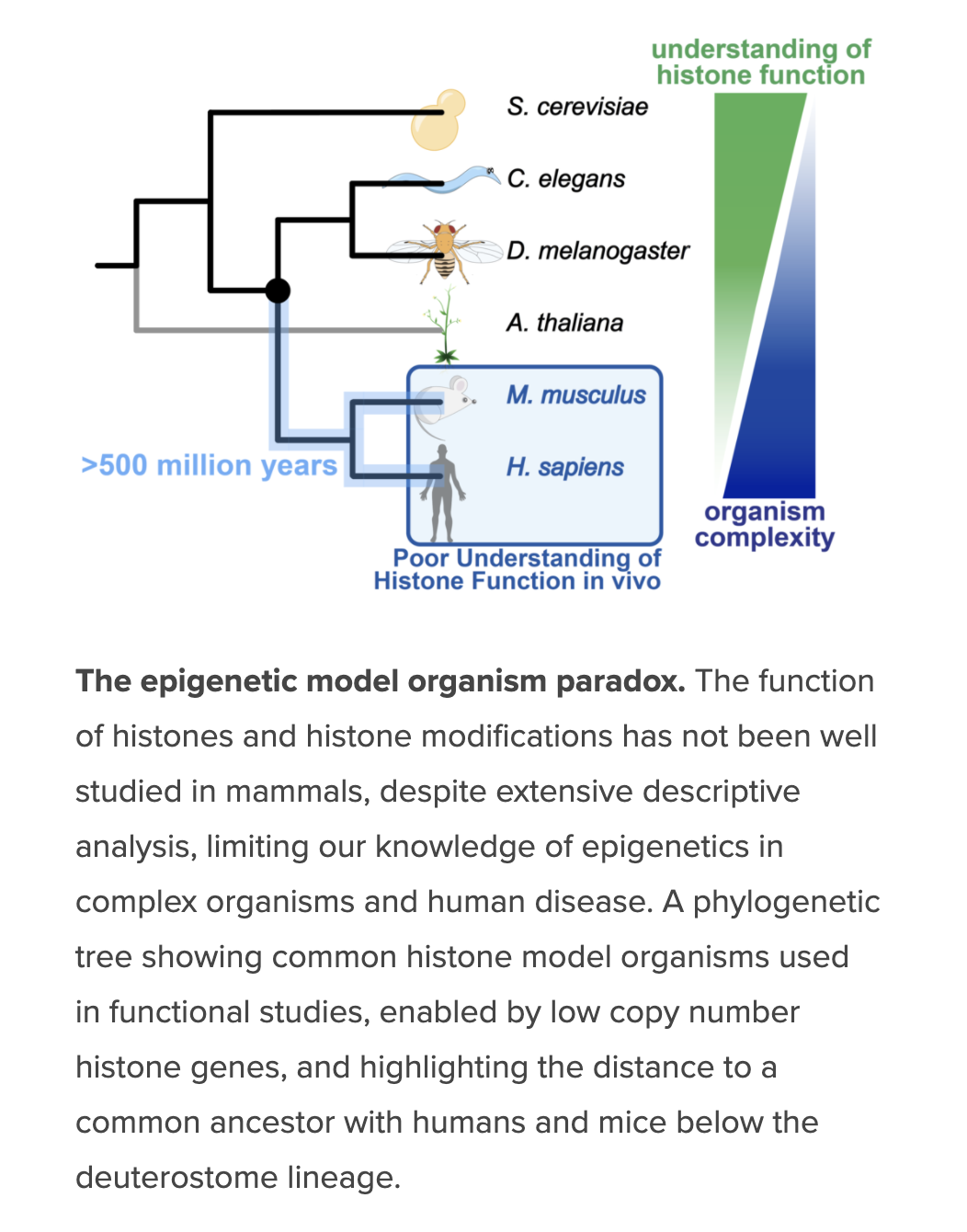
Despite widely held assumptions and abundant descriptive data, we lack an understanding of the function of histone modifications in complex organisms, i.e. deuterostomes, vertebrates. This is due to the experimental intractability of histone genetic complexity. A motivating challenge for the lab has been to address this fundamental gap in the field of epigenetics, which is central to understanding how large genomes in complex organisms are regulated to direct cellular differentiation and rapid environmental responses across physiologic systems and their dysregulation in disease. We are building a platform for functional histone studies in mammalian systems. With these approaches, we set out to address fundamental questions: What are the roles of histones and their modification in mammalian cell differentiation in vivo? Do individual histone residues and their modification have cell type specific effects, instruct cell fate, or direct malignancy? Are some essential for all cell types?
A long term goal is to apply mechanistic knowledge of epigenetic regulation and functional histone genetic tools to understand epigenetic processes in immune cell development and function including immunologic memory, trained immunity, immune cell exhaustion, and more.